Background
With the increase in using patients’ own cells (e.g., stem cells, immune cells) as a living drug to cure disease, large-scale cell manufacturing has become a new form of pharmaceutical manufacturing. Using process analytical technology (PAT) to control the manufacturing process to meet FDA requirements is critical. For cell manufacturing, the PAT technology to provide online real-time monitoring of cell density, morphology, and their changes is important. However, there is currently no fully automatic technology to measure the cell changes in situ during the cell manufacturing process.
Our previous success in cellular-level computer vision applications has proven our ability to combine external domain expertise from the world of biology and pathology with our own expertise in computer vision and machine learning. Thus, a primary objective of this exploratory research was to determine the feasibility of developing a low-cost cell imaging system, without the need for cell staining, suitable for bedside cell therapy. In addition, an automated approach to detect the cells of interest was needed for further automated cell characterization.

Figure 1: Visual feature collection apparatus components.

Figure 2: Complete visual feature collection apparatus system.
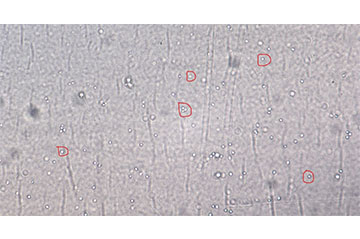
Figure 3: Label-free cells of interest
Approach
 Figure 4: Automated cell detection. Left: An extracted H&E patch showing tumor cells embedded in stromal tissue with tumor-invading lymphocytes. Right: Network posterior map of lymphocyte locations.
Figure 4: Automated cell detection. Left: An extracted H&E patch showing tumor cells embedded in stromal tissue with tumor-invading lymphocytes. Right: Network posterior map of lymphocyte locations. Development of the cell imaging system, or the Visual Feature Collection Apparatus, was the first step of this process. Since purchasing an off-the-self, benchtop microscope system would not be suitable, the team selected individual microscope components (lighting, objective, camera, etc.) (Figure 1) to create a more compact version of a typical benchtop microscope system. To aid in the component selection process, we tested the imaging capabilities of the composite system by imaging microspheres 10 µm in diameter. Since the microspheres are similar in size and transparency to the cells of interest, they allowed us to more efficiently identify the appropriate components without having to wait for cell cultures to be populated. After the Visual Feature Collection Apparatus was finalized (Figure 2), we collected samples of Peripheral Blood Mononuclear Cells (PBMC) from a flow-based bioreactor to generate images to show the cell features for automated cell detection (Figure 3). The task of developing an automated cell detection model was performed in parallel to the Visual Feature Collection Apparatus task. Our team identified a publicly available dataset to serve as a proxy for live-cell imaging. The dataset contained image patches extracted from whole slide images of hematoxylin and eosin (H&E) stained pathologic sections of breast tissue scanned at 20X magnification providing location annotations of lymphocytes, malignant epithelial, and normal epithelial cell nuclei. With this data, a fully convolutional regression model was designed to identify instances of lymphocytes by predicting a posterior map of cell center likelihood, thus enabling the detection of the cells of interest (Figure 4).
Accomplishments
A low-cost Visual Feature Collection Apparatus was developed, suitable for application of bedside cell therapy was developed. In addition, with the combined expertise of domain subject matter experts in biology and pathology, our team demonstrated the feasibility to image live cells, without cell staining, using a low-cost piecewise imaging system, in addition to the capability of detecting cells of interest. Furthermore, our results have positioned us for additional research of cell quantification and characterization.
