Pathology is a critical part of screening, detecting and staging cancer and other diseases, and yet the number of board-certified pathologists has been declining in recent years.
DETAIL
An algorithm is a set of steps for a computer program to accomplish a task.
Over the long term, the demand for disease detection in an aging population has increased pathologist workloads. In the short term, this growing backlog of work is surging as people who put off cancer screening and other health diagnostics due to the COVID-19 pandemic are once again seeking diagnostic health care services.
These trends are helping to increase research and development of artificial intelligence and other technologies that can speed up analysis of cells for a variety of diseases. Southwest Research Institute is collaborating with several local research institutions and physicians to develop machine vision algorithms that will increase the speed and accuracy of cancer detection and other diagnoses.
ABOUT THE AUTHOR
Hakima Ibaroudene launched and leads the Medical Diagnostic and Prognostic Technologies program area in SwRI’s Intelligent Systems Division. The program applies artificial intelligence to solve problems in the medical domain to address challenges in imaging, image processing and electronic health record data. Ibaroudene currently leads research in mental well-being, pathology, radiology, biomechanics and cognitive performance.
Our work in this space began in 2018, over a year before the onset of the COVID pandemic, when we trained algorithms that had been used in perception systems for industrial robotics and automated vehicles to detect breast cancer from digital pathology slides. The SwRI research with UT Health San Antonio pathologists placed first in the BreastPathQ: Cancer Cellularity Challenge conducted by the American Association of Physicists in Medicine, the National Cancer Institute and SPIE, the international society for optics and photonics.
DETAIL
Digital pathology uses cell imagery from slides scanned into a computer for analysis by a pathologist. Pathologists are physicians who use microscopes and other instruments to study tissues and cells to identify abnormalities and disease. They typically obtain cells via a biopsy or surgery. The process can take several hours to positively identify cancer from a single sample and multiple samples are often required.
By winning that worldwide breast cancer detection competition, the Medical Diagnostic and Prognostic Technologies program at SwRI quickly gained recognition in the world of digital pathology research, but that was only the beginning of a rapidly growing program area and ongoing collaboration with local medical professionals.
DIGITAL DETECTION TRAINING
Algorithms are used in cancer research, especially in the fields of diagnostics and prognostics, using digital image data to increase the speed and efficiency of pathology, while also providing data that can be mined for future treatment options.
Having never worked with medical data or tissue images, SwRI engineers seemingly were at a disadvantage in comparison to the competition. However, they were confident that algorithms for autonomous robots could be adapted to new applications. The team worked with pathologists Dr. Bradley Brimhall and Dr. Edward Medina at UT Health to develop digital diagnostic strategies. The pathologists provided labeled digital files distinguishing between cancer cells and normal cells in hematoxylin and eosin (H&E) stained slides of tissue samples. Then computer scientists set out to automate the process using artificial intelligence techniques, specifically using machine learning.
DETAIL
Hematoxylin and eosin (H&E) stain is one of the most widely used tissue treatments for histology, which studies the microscopic anatomy of tissues taken during a biopsy of a suspected cancer.
Out of 87 competitors from some of the world’s top research institutions, SwRI’s algorithm most closely matched the ground truth diagnoses by physicians. With this initial success in applying machine learning to digital pathology, SwRI was eager to tackle more problems in the medical space. First, engineers stepped back and considered the purpose of research efforts. In this case, even the most qualified pathologists are subject to human error. Accuracy can vary depending on factors such as sleep deprivation and clinician experience. Because pathologists perform many other diagnostic, prognostic and research tasks throughout their day, automating one of the most time-consuming tasks allows them to devote more time to advancing cancer treatment in other ways.
MACHINE LEARNING
The team used deep learning techniques, also known as convolutional neural networks, to adapt algorithms to learn from datasets of cancerous and normal cells. These machine learning algorithms automate the processing and interpret large amounts of complex data by extracting and learning patterns.
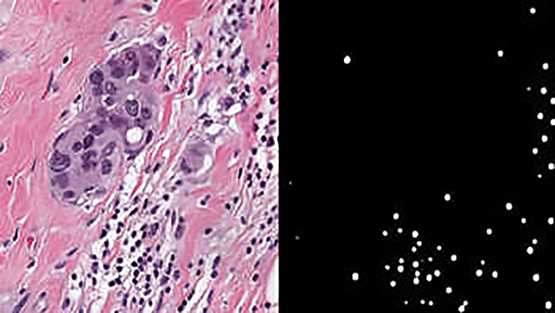
SwRI developed a detection algorithm using breast cancer tumor cell images to compete in BreastPathQ: Cancer Cellularity Challenge. Out of 100 submissions, the SwRI solution placed first in the international challenge to develop an automated method to detect breast cancer tumor cells.

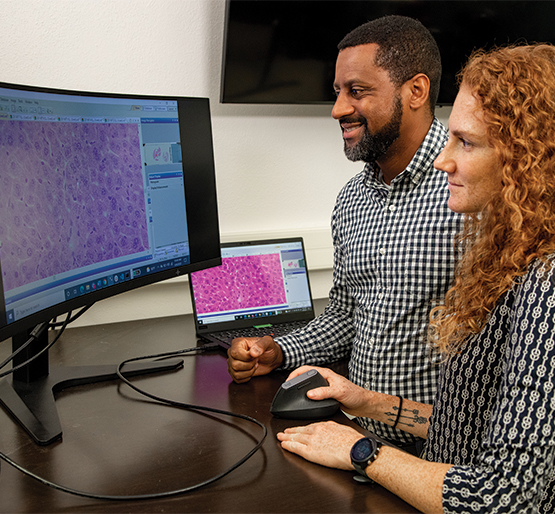
SwRI analysts David Chambers (left) and Hakima Ibaroudene (right) work with Dr. Bradley Brimhall of UT Health on an AI tool that analyzes stained pathology slides. The purple shapes are irregular blood cells known as follicular lymphoma (FL), a B-cell malignancy, and Philadelphia chromosome (Ph)-negative.
DETAIL
Artificial neural networks are computing systems inspired by biological neural networks in animal brains. Convolutional neural networks are most commonly used to analyze visual imagery.
The team used an artificial neural network (ANN) as a predictive model to classify data. Known inputs are fed into an ANN’s top layer, which passes those values through one or more hidden layers. The supervised learning algorithm analyzes the training data and produces an inferred function, which can be used for mapping new examples. An optimal scenario will allow for the algorithm to correctly determine the class labels for new, unlabeled datasets. This requires the learning algorithm to generalize from the training data in a “reasonable” way. ANNs have a high tolerance for noisy data and excel at classifying patterns.
An image recognition neural network breaks down and examines different features. The algorithm is optimized through trial and error and computational back propagation through the various layers of the neural network, eventually narrowing down its predictions to something that is accurate. The learning process takes the inputs and the desired outputs and updates its internal state accordingly, so the calculated output is as close as possible to the desired output.
Advancing Digital Diagnostics
One key to advancing digital diagnostic capabilities is developing and maintaining relationships with collaborators in the medical field. SwRI engineers recognize the importance of consulting medical professionals for their needs and expertise. Since winning the BreastPathQ competition, SwRI has completed projects in collaboration with various local medical professionals. For instance, together with Brimhall and Medina, SwRI developed an algorithm to determine the hormone receptor status of breast cancer cells.
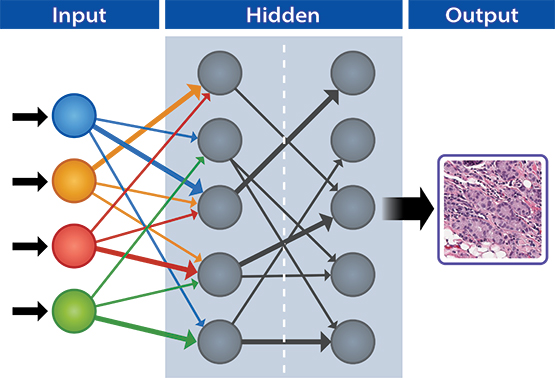
Convolutional neural networks can be trained to detect features in images. Data are entered at the input layer (far left column) and analyzed in various hidden layers (middle columns) where images are compared to identify cancer or other anomalies based on machine learning inputs. The output layer (right column) identifies images that have cancer cells.
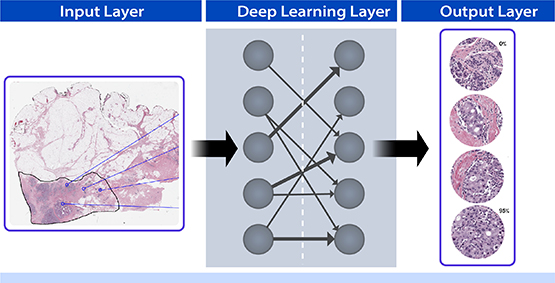
Left column: An image of a pathology slide (breast tissue with H&E stain) is digitized and input into a convolutional neural network to train it to differentiate between normal and cancer cells. Most breast cancers arise from epithelial cells (stained purple) that normally line the lobules and ducts that produce milk. Middle column: Hidden layers of a neural net use deep learning algorithms to compare input images to thousands of similar images to teach the tool to identify cancer cells. The neural net uses contextual information to identify epithelial cells where they should not occur or arranged in abnormal patterns, indicating cancer. Right column: Training a deep learning algorithm to identify cells with and without cancer improves the accuracy of cancer cell detection. The output layer indicates the percentage of cancerous cells in a sample, depending on the collection site or configuration.
DETAIL
The Allred score was named for the doctor who developed the technique for assessing a cancer’s hormone receptor status. The technique combines the percentage of positive cells and their intensity to determine the Allred score. Scores from zero to two are considered negative. Scores from three to eight are considered positive and would likely respond to hormone therapies.
Hormone receptor status allows doctors to better determine an effective treatment plan, particularly if a cancer is likely to respond to hormonal therapy. However, the current method is susceptible to interpretation and human error, which could be eliminated by automation. This project used estrogen receptor (ER) immunohistochemistry (IHC) staining assay, rather than H&E stains, which presented new challenges. The staining process makes hormone receptors show up in a sample of breast cancer tissue. The assay first identifies a percentage of cells out of 100 that are positive for hormone receptors and their intensity, or how well the receptors show up after staining. The intensity corresponds to how susceptible the cells are to hormonal influences. Because ER IHC slides are drastically different than IHC slides, engineers had to take a new approach that included hand-labeling cells and assessing pixel intensities. This information is then combined to score the sample on a scale from 0 to 8. The higher this “Allred” score, the more receptive the cancer will likely be to hormone therapy.
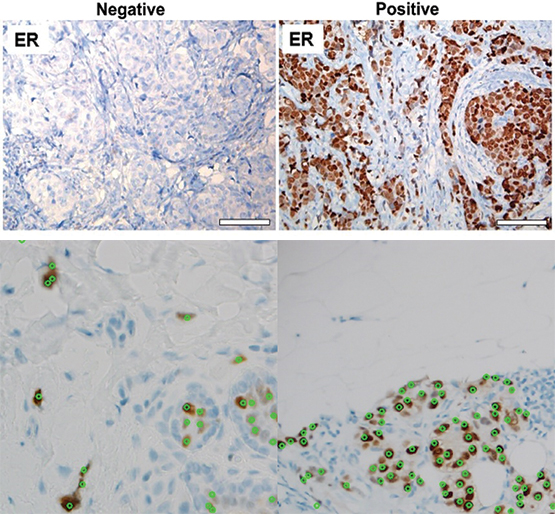
SwRI algorithms accurately determined the hormone receptor status of biopsies at top. Below, the program identified hormone receptor cells (green) while indicating false positives (red) to help pathologists determine if a cancer will respond to hormonal therapy.
SwRI’s Daniel Poole and Dr. Courtney Rouse adapt machine learning algorithms to identify polyploid cells in DLBCL samples to help clinicians assess new polyploid suppression therapies. Chemotherapy-induced polyploid cells become resistant to additional bouts of chemotherapy.
Predicting Drug Response
To expand our expertise beyond breast cancers, SwRI needed to expand its network of medical collaborators. The group contacted Dr. Daruka Mahadevan of Mays Cancer Center (part of UT Health San Antonio) after reading an article about his lab’s recent drug discovery research for one of the deadliest forms of cancer, Diffuse Large B-Cell Lymphoma (DLBCL).
DETAIL
Polyploidy means that cells of an organism have more than one pair of chromosomes. Most species whose cells have nuclei are diploid, meaning they have two sets of chromosomes. Polyploidy may occur due to abnormal cell division.
Mahadevan has new evidence that DLBCL becomes resistant to treatment as chemotherapy results in the formation of large polyploid cells with more chromosomes than found in healthy cells. To address this issue, Mahadevan tested the effects of other drugs administered alongside chemotherapy to resist the development of these problematic cells. Unfortunately, there’s no methodology for efficiently determining how effective a drug was at eliminating polyploidy. Moreover, because treatment for polyploidy is not yet approved, most pathologists are not trained to identify these cells. Like many other digital pathology projects, the motivation behind automating this process is to eliminate human error and accelerate discovery of therapeutics. A machine learning algorithm was first trained and tested on images of cultured polyploid and normal diploid cells. Currently, the team is adjusting the algorithm to assess images of tissue data.
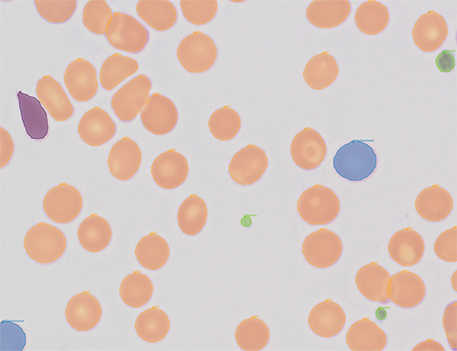
SwRI created an AI tool that analyzes stained pathology slides, looking for irregular blood cells that indicate lymphoma.
Advancing Tools, Capabilities
Throughout their time working on digital pathology research, SwRI engineers have built an image labeling tool that can be used for a variety of data. Project-specific modules can be added as needed, making the tool incredibly versatile, even outside of medical applications. Each project has required a new skillset, which has expanded the capabilities of the group as a whole, enabling the team to apply them to new challenges.
While some of these projects are ongoing, the members of the medical diagnostic and prognostic program area continue to apply for funding, brainstorm with current collaborators and reach out to potential new collaborators. With so many types of cancer and disease, the experience gained from current and past projects will have countless future applications. The group is passionate about improving cancer treatment by developing tools that doctors find useful, timesaving and cost-effective.
Questions about this story or Bioinformatics Data Analysis Services? Contact Hakima Ibaroudene at +1 210 522 3963.

