Background
As of 2016, breast cancer is the most commonly diagnosed cancer in females, with over 200,000 new cases annually since 2006. With diagnostic rates of this magnitude, there is a significant need for improved digital pathology tools to assist and automate parts of the traditional pathology workflow. Breast cancer is traditionally diagnosed by pathologists using Hematoxylin and Eosin (H&E) stain and assessed visually for morphology. Once morphology is assessed, tissues are stained using immunohistochemistry (IHC) to evaluate predictive biomarkers, such as hormone receptor presence. Unfortunately, these tests introduce human error in both application and assessment. This project’s purpose was to research the feasibility of using SwRI’s existing neural network expertise to classify breast cancer into different groups using hormone receptor status.
Approach
Using a small dataset of estrogen receptor (ER) IHC stained tissue, we hand-labeled over 10,000 individual stained cells using an SwRI-developed labeling tool. Each cell was labeled and the pixel location recorded for post-processing and network training. We developed a custom data layer to allow the network to accept this data. We then trained with the Inception version 3 (v3) backbone, a common image recognition model, encoding keypoints at a stride of 8 with non-normalized Gaussian distributions with standard deviations of 1.75 (units of pixels x 8). This network was optimized against a Euclidean loss layer and trained for 3,000 iterations using the Adam optimization algorithm, an extension of the traditional gradient descent methodology that is popular within the deep learning computer vision space. The data was augmented at training time with scale, hue/intensity shift, and 360° rotation.
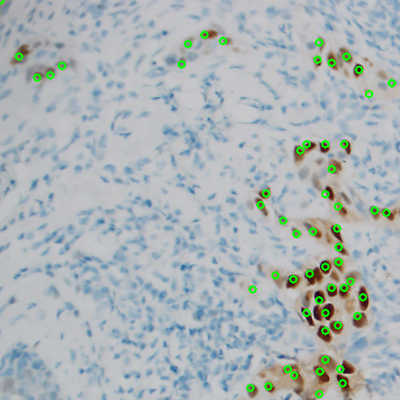 Figure 1: ER stained cells identified by the neural network, with green circles representing correctly identified cells.
Figure 1: ER stained cells identified by the neural network, with green circles representing correctly identified cells. Accomplishments
The trained neural network was able to correctly identify individual cells in ER stained tissue as shown in Figure 1. Evaluation of precision recall and associated debug images show that in some images, especially those with lower magnification, more thorough labeling is required. The network is correctly labeling cells that are being classified as false positives because the original slide was under-labeled. The network was outperforming the hand labeled data. A larger, curated dataset would be necessary and will be used in future research to map these results to actionable data for pathologists.
