The search for solutions to human suffering dates back to the beginnings of civilization. The earliest treatments were typically based on observations of nature and superstitions, consisting largely of some combination of ritual and plant- or animal-based ingredients. Modern drug discovery began in the 1800s, when scientists began to isolate compounds from natural sources to create the first drugs. The 1900s saw advancements in synthetic chemistry, antibiotics and an understanding of drug mechanics.
In the early 2000s, the onerous drug discovery process developed in the 20th century remained largely unchanged. While scientists no longer manually screened thousands of compounds in search of viable drug candidates, high-throughput screening methods developed in the 1990s only automated screening for a few hundred to a few thousand compounds at a time.
Developing viable candidates into a clinical drug remained a 10- to 15-year endeavor. For context, Alexander Fleming’s 1928 discovery of penicillin did not start saving patient lives until the 1940s. The cancer-fighting drug Keytruda®, developed in the early part of the 21st century, took about as long to arrive on pharmacy shelves in 2014.
Around 2010, Southwest Research Institute tapped its multidisciplinary expertise in computer science and chemistry to develop a rudimentary version of Rhodium™, SwRI’s proprietary molecular docking platform.

This Rhodium image shows how various small-molecule keys bind with a large, disease-causing molecule, allowing chemists to assess the best drug candidates for halting or treating infections.
A grandfather clock offers an apt analogy for drug discovery that connects the contexts for antivirals or cancer therapeutics. The clock represents a large disease-causing molecule such as a protein produced by a virus or tumor needed to spread or grow. The protein requires special keys, or small molecules, that fit precisely into a keyhole or binding site to cause undesirable replication. For a clock, the key winds the mechanism that keeps it running. Viruses or invasive tumors will spread or grow unless the replicating agent is locked out. In the context of antivirals, drug developers look for “inhibitor” keys or ways to stop the “clock” and thwart infected cells. These inhibitors may compare to tipping the clock over, jamming its hands or filling in the keyhole. Rhodium identifies inhibitors to stop the clock and ranks how well those small molecules will perform against viruses or, for oncology, address rapid, uncontrolled cell growth.
Rhodium software enables computer-aided drug design and structure-based virtual screening to accelerate drug discovery and allow for strategic development of pharmaceuticals.
Molecular docking uses computational methods to predict the preferred orientation of one molecule to another, like a ligand binding to a protein. It essentially predicts how a small molecule, the ligand, will meld with the binding site of a larger molecule, the protein, similar to a lock and key.
Scientists began using Rhodium for SwRI’s medical countermeasures research to speed up the search for viable antidotes for nerve agents and pesticide exposure. With Rhodium, picking winning compounds was no longer a lengthy process of elimination nor a matter of chance. The initial code for Rhodium provided a springboard for further development.
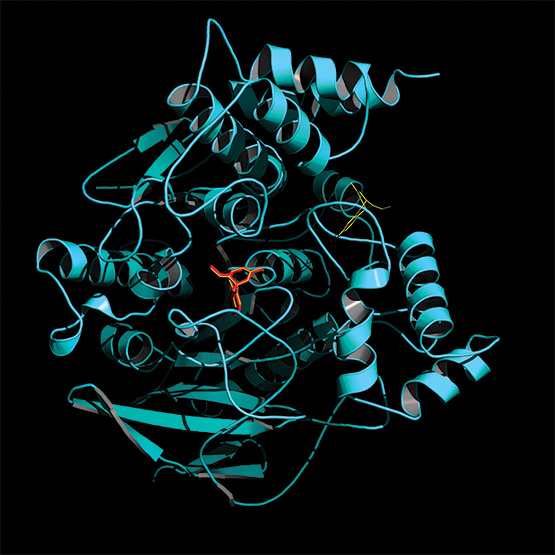
In 2013, chemists used Rhodium software to screen potential treatments for Alzheimer’s disease (red), shown here interacting with a neural enzyme (cyan).

In 2017, SwRI tapped into mobile communications technology to turbocharge Rhodium software, accelerating its processing capabilities. The resulting optimized “supercomputer” enabled streaming up to four times faster to visualize in 3D how an organic structure captures a powerful anesthetic.
Rhodium Edge
DETAIL
A protein crystal structure depicts the three-dimensional structure of molecules, determined through crystallography, that reveals the protein’s atomic structure.
Rhodium pinpoints and predicts precisely where a drug will likely bind to a protein associated with a disease based on its 3D structure. When a drug molecule binds with a protein, it can block or modify the functionality of the protein to circumvent disease.
SwRI’s pharmaceutical and bioengineering researchers support every facet of drug discovery and development through clinical trials, following Current Good Manufacturing Practices. Rhodium molecular docking services supercharge what’s possible for clients, allowing comprehensive 3D analysis of the protein crystal structure of target molecules, such as proteins key to diseases. It is particularly advantageous during the initial drug discovery phase, because conducting early screening for new medications is expensive, time consuming, and may be limited by specialized lab access, such as biosafety constraints in discovery of new anti-infective drugs. Eventually new drugs are tested in clinical trials. But first, meeting strict potency and purity standards is required for obtaining FDA approval for a trial.
DETAIL
In silico means in or on a computer, referring to the silicon often used to make computer chips.
Rhodium can narrow down the list of potential treatments for disease, scouring massive in silico compound libraries for those with a high probability of effectiveness or potency, using fully traceable simulations. SwRI offers small molecule protein docking simulations as a service to SwRI clients and collaborators to support their drug development programs.
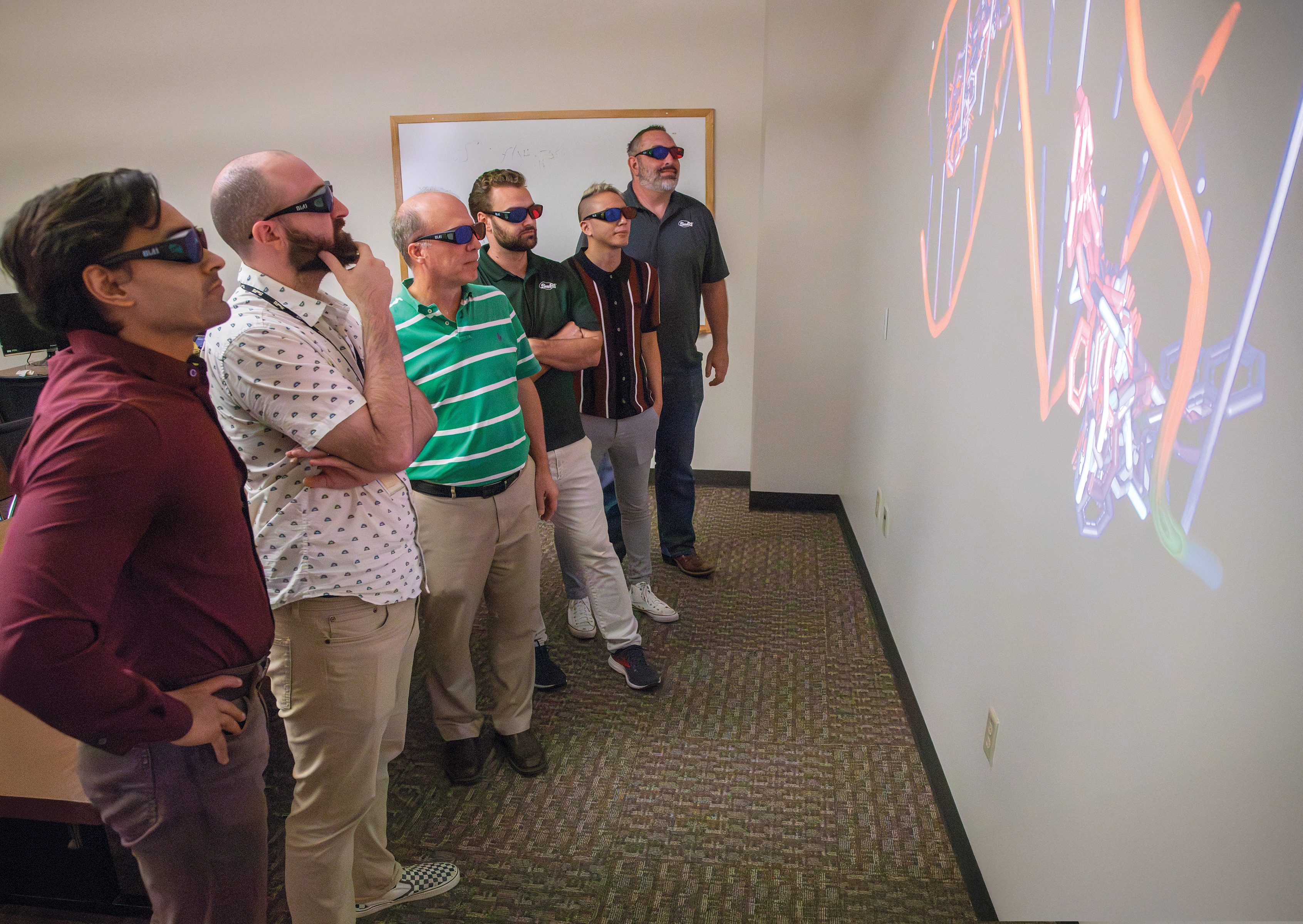
SwRI’s drug discovery team wears 3D glasses to visualize the protein crystal structure of diseases in three dimensions to understand how an inhibitor compound may bind to the disease to prevent spread. The 3D analysis of the disease structure is performed using Rhodium software.
A Decade of Development
In 2014, Rhodium could screen around 25,000 compounds a day. Even those early iterations were a breakthrough for scientists. Through the integration of machine learning, graphical processing and expanded computer capacity over the last decade, Rhodium can now screen 250,000-plus compounds each day, increasing the number of candidates screened and the likelihood of success. A direct-search grid pattern algorithm delivers a “first-pass” solution in one-tenth of a second, which is subsequently refined to provide accurate and speedy results, making it possible to find the proverbial needle in a haystack.
The software also complements the development of laboratory assays, procedures that measure the biological effect of a drug candidate in a research setting. In connection, Rhodium can help assess compounds as control drugs or tools to study diseases. Virtual screening also saves on capital equipment and laboratory space, which is of particular concern when exploring treatments for highly infectious diseases or antidotes for toxic nerve agents, where testing is limited by biosafety constraints.
COVID’s Wake
Somehow the COVID-19 pandemic simultaneously feels as if it happened in the distant past and much more recently. More than five years ago, SwRI joined worldwide efforts to rapidly find coronavirus cures to save lives. First, SwRI’s drug discovery and development team increased Rhodium’s processing power to evaluate treatments. Using internal research funding, the team expanded the software, allowing it to take advantage of high-performance supercomputers to screen 40 million drug compounds based on the structure of the Sars-CoV-2 virus main protein. SwRI continues to use Rhodium to search for broad-spectrum antiviral treatments, prepare for future pandemics and support other medical countermeasures. In 2024, SwRI received two patents for inhibitors to target coronavirus spike receptors for SARS-CoV-1, SARS-CoV-2, MERS-CoV and other variants and antiviral drugs designed to target the N-terminal domain, found at the beginning of the protein chain, for the spike receptor binding domain of the coronavirus. Additional research continues for antiviral drugs treating Dengue virus and Lassa fever.
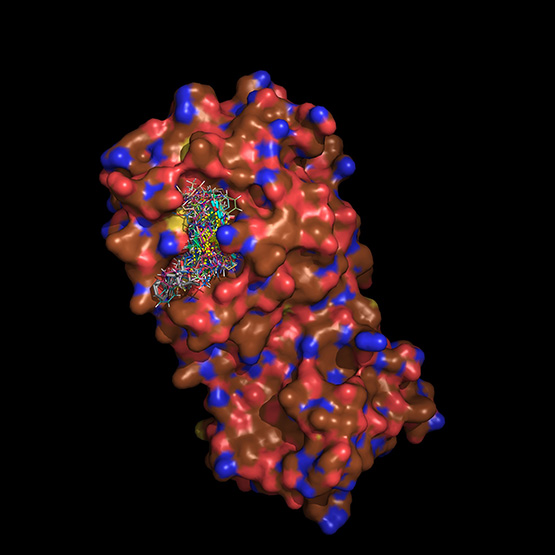
Courtesy of SwRI/Protein Databank (rcsb.org), Entry 6LU7
SwRI’s Rhodium virtual screening software used this 3D model of a SARS-CoV-2 protein to evaluate millions of drug compounds to identify therapies effective against COVID-19.
SwRI scientists used Rhodium’s molecular docking platform to predict the DNA binding affinity and cell toxicity of cancer-fighting drugs.
DNA Domain
In 2024, SwRI expanded Rhodium’s capabilities beyond proteins to screen DNA-targeting compounds and visualize and rapidly predict how DNA-targeting therapeutics can attack cancer cells as well as other diseases. While a number of drug development platforms and machine learning methods virtually screen drugs that target proteins, fewer methods exist for screening drugs that target DNA.
According to the World Health Organization, cancer is a leading cause of death worldwide, responsible for one in six deaths globally. Chemotherapy uses a combination of drugs to slow or stop the growth and spread of cancer cells and shrink tumors. Many of these chemotherapeutics directly target DNA, slowing cancer growth but potentially damaging the DNA in healthy cells and causing severe side effects, medical complications and even secondary metastases. Cancer cells often have mutated DNA repair machinery and replicate much faster than healthy cells, which makes DNA a viable target for selective cancer treatment.
SwRI has now successfully demonstrated a virtual screening application to design more selective DNA-targeting therapeutics to combat different types of cancer and infectious diseases. Researchers virtually screened numerous cancer-fighting compounds, ranked their potential effectiveness as treatments and are now applying the software to design next-generation chemotherapies.
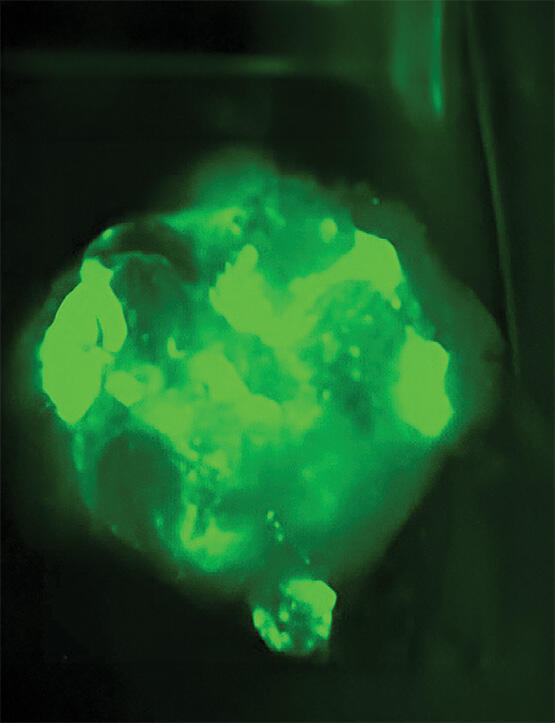
Courtesy of Texas Biomedical Research Institute
Researchers at SwRI, UTSA and Texas Biomed used SwRI’s Rhodium software to identify viable compounds to treat emerging zoonotic pathogens, such as this Nipah virus infection growing in this humanized organoid. These emerging viral threats can jump from animals to humans, where they are particularly lethal.
Measles Mission
SwRI collaborated with The University of Texas at San Antonio, Texas Biomedical Research Institute and the UT Health Science Center to identify more than two dozen viable treatments for zoonotic pathogens that can jump from animal hosts to infect humans. The team used Rhodium software to map the protein structure of the measles virus, searching for a broad-spectrum treatment that could potentially treat Hendra and Nipah viruses. The measles virus is closely related to these deadly bat-borne henipaviruses, which are endemic to some parts of the world and cause particularly lethal infections in humans.
Researchers are particularly interested in these viruses because they frequently spillover from animals to humans, which elevates their pandemic potential. Out of 40 million compounds, Rhodium identified 30 potentially viable viral inhibitors that attach to a protein present on henipaviruses. Virtual screening narrows the list of potential treatments while reducing demands for high-containment laboratories and saving time and resources. More research is needed to validate and develop the viral inhibitors Rhodium identified. In early 2025, a measles outbreak in Texas highlighted the need for broad-spectrum treatments.
DETAIL
A zoonotic disease is transmitted between animal and human hosts. These infections are caused by pathogens, such as bacteria, viruses, parasites or fungi.
Playing Games
In 2025, SwRI continued its expansion of the Rhodium platform with the Generative Approaches for Molecular Encodings (GAMES) model. Through multidisciplinary research, SwRI’s drug discovery experts collaborated with Institute engineers specializing in artificial intelligence to develop the model. GAMES generates valid Simplified Molecular Input Line Entry System (SMILES) characters, which are text-based language encodings for candidate drugs. Researchers trained the GAMES model to understand and generate accurate new combinations of SMILES characters. SwRI scientists use the SMILES language to encode this information for use by Rhodium.

Rhodium accelerates the drug development process by narrowing down the best candidates for conventional development and evaluation processes.
SwRI’s LAMP initiative, an internal research program focused on the adoption and advancement of large language models (LLMs), funded the program. Using LLMs with drug discovery and generative artificial intelligence offers the potential to deliver an almost infinite number of new treatments. These candidates could be synthesized to produce viable drug candidates.
GAMES demonstrates a systematic way to build databases of molecules for AI processing and comparison using a language-based method. This offers advantages over traditional data science methods such as Quantitative Structure Activity Relationships (QSARs). Developed in the 1960s, QSARs use linear mathematical models to connect chemical structures with biological properties, relying on mathematical representations called descriptors or fingerprints. Mathematical and graphical representations are already integrated into Rhodium. Incorporating non-linear GAMES into the Rhodium workflow offers a way to go beyond traditional descriptors to explore the generative possibilities available through large language models.

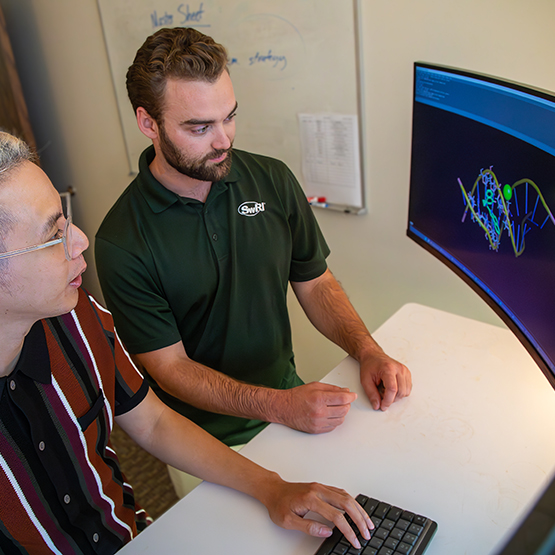
SwRI developed a large language model known as GAMES to generate Simplified Molecular Input Line Entry System (SMILES) strings. The lower text-based string represents the structure of the chemical molecule above.

SwRI developed the GAMES machine-learning application to generate valid SMILES characters (background), text-based language encodings for candidate drugs. GAMES provides a systematic technique for building databases of molecules, represented by the chemical structure in the foreground, for AI processing within the Rhodium platform.
Timeline
2011 — Rhodium software development begins
2012 — Internal research program adds GPU processing capability
2015 — Grand Challenge docking contests demonstrate accuracy
2020 — Rhodium used to screen for drug treatments for coronavirus
and other infectious diseases
2023 — Machine learning integrated into Rhodium software
2024 — Nipah and Hendra antiviral poster presented in Australia
2025 — GAMES model created

ABOUT THE AUTHOR
Dr. Jonathan Bohmann is a staff scientist who pioneered the development of SwRI’s high-performance Rhodium molecular docking platform. This novel simulation program has proven applications in diverse areas of drug discovery, including neuroscience, virology, oncology and anti-infectives. Bohmann presented a poster about research into henipavirus countermeasures at the Hendra@30 conference sponsored by Australia’s National Science Agency CSIRO. Bohmann co-authored the work with Dr. Stanton McHardy of The University of Texas at San Antonio and Dr. Olena Shtanko of Texas Biomedical Research Institute.
2025 Forward
Accuracy, selectivity and proven results all top the list of reasons Rhodium continues to play an integral part in SwRI’s comprehensive drug discovery and development process. With its world-class computer-aided drug design and virtual screening, Rhodium software provides fully traceable simulations and comprehensive 3D protein crystal structure analysis to continuously push new therapeutics forward. Rhodium’s revolutionary capabilities have enabled numerous pilot projects and large multiyear research programs. Over the next 10 years, SwRI hopes to continue to discover new and viable pharmaceuticals and biologics key for treating cancer and infectious diseases while anticipating how to conquer the next possible pandemic. SwRI’s Rhodium software is a 21st century solution for accelerating drug development with the ultimate goal of alleviating suffering and saving lives.
Questions about this story or Rhodium Molecular Docking Software? Contact Dr. Jonathan Bohmann at +1 210 522 5219.
