Rhodium™ is a moleculardocking software tool developed by chemists and computer scientists at Southwest Research Institute. Rhodium software as a service (SaaS) is available to clients from industry, government and academia. This virtual screening software integrates graphical processing, software and Machine Learning in Drug Discovery to scan hundreds of thousands of drug compounds per day for Structure-Based Drug Design projects.
Protein Docking Software for Drug Design
Rhodium’s high throughput 3D analysis of protein docking helps scientists select ligands to understand how drug compounds might interact with proteins. The Rhodium executable is a command line interface (CLI) platform, enabling us to perform continuous development to ship new integrated algorithms as the field evolves and hardware advances. Protein docking software for drug design features include:
- Unbiased comprehensive in silico docking on protein structures
- Extremely high throughput
- Demonstrated efficacy in multiple druggable targets
- Superior selectivity for selection of true ligands
- Superior accuracy in predicting binding poses
- Ideal for allosteric sites and protein-protein interactions
Download our white paper to learn how Rhodium is a top performer against other molecular docking platforms or contact Jonathan Bohmann, Ph.D. for more information.
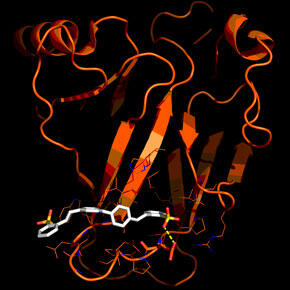
Rhodium™ Molecular Docking Software
Learn how SwRI trained its Rhodium Molecular Docking Software to ensure accuracy when evaluating millions of potential drug compounds.
Rhodium Direct Search Docking Algorithm
The numerical method implemented in C/C++ is a direct search algorithm used to find the maximum or minimum of an objective function, i.e. the elements of protein crystallographic coordinates interacting with a ligand. In the case of Drug Discovery Research, this is considered as a non-linear optimization problem. The core function is written in assembly to enable direct resource allocation that achieves optimized computing performance of rigid-body atomistic simulations. The direct search algorithm can be visualized in a graphical output from the electromechanical pen plotter software package called from CalComp programs.
SwRI-developed Rhodium™ scans compounds for the ability to bind and inhibit a target; in this case, the software is optimizing the unbiased docking of a potential treatment based on the protein structure of the Nipah virus, which is closely related to the Measles protein structure. The video has been sped up to enhance visualization.
Visit Machine Learning in Drug Discovery to learn more about our molecular docking scoring algorithm or contact Jonathan Bohmann, Ph.D. for more information.
Pharmacophore Modeling
In the absence of detailed protein crystal structure data, small but rich bioactivity datasets of an analog series can provide valuable insights for lead optimization and early drug discovery projects. The pharmacophore is an abstract mathematical representation of key atom features. The common features are interpreted as pharmacologically relevant concepts that guide the design process. This pharmacophore modeling approach creates a structured framework within the drug design space to reduce complex data and enhance the precision and efficiency of our drug development efforts.
| Common Feature | Chemical Description | Atom Type Examples |
| HBA and HBD | Hydrogen-bond acceptor and hydrogen-bond donors | Brönsted acids and Lewis bases |
| PIC | Positive ionizable center | Amines and sp3 hybridized nitrogen |
| HYDROPHOBIC | Lipophilic and “greasy” | sp3 hybridized carbons |
| HBD | Hydrogen-bond donor | Brönsted acids |
| HBA | Hydrogen-bond acceptor | Lewis bases |
| AROMATIC | Planar & cyclic rings | sp2 hybridized carbon, nitrogen, oxygen and sulfur |
| POLAR | Ions | Any non-covalently bonded ionic species |
| ANION | Negative ionizable centers | Oxyanions, carboxylates and enolates |
Molecular Visualization Studio
Effective structure-based virtual screening relies on sophisticated molecular graphics visualization tools. Our department is equipped with a state-of-the-art visualization studio, designed for collaborative engagements with clients, informally dubbed the “Protein Cave.” The Modeling Lab studio allows us to vividly present and discuss research findings, enhancing our ability to model and visualize protein-ligand complexes. Through molecular docking software and ligand-based pharmacophore modeling, we provide detailed insights into molecular interactions, ensuring that our structure-based virtual screening processes are both precise and informative.

