BACKGROUND
Amino acid detection is a driving science requirement for NASA and is the scope of current and future flight missions to Mars, Europa, Enceladus and Titan. The search for life outside our planet and throughout our solar system is fundamentally determined by the presence of amino acids and the ratio of their chiral enantiomers. There exists a multitude of methodologies for laboratory based amino acid extraction and detections. However, these methods are impractical in harsh and remote locations such as outer planets. There exists a need for a simple sample preparation process that minimizes the need for introduced chemicals/solvents and also minimizes the need for sample transfer between additional vessels.
APPROACH
To meet this challenge, a single-step process for the removal and derivatization of amino acids using a solid-phase-micro extraction (SPME) technique followed by in-situ derivatization was explored. The single-step exposure, removal and derivatization process would be followed by immediate sample analysis on a gas chromatograph coupled to a mass spectrometer (GCMS). A carboxen/divinylbenzene/poly dimethyl siloxane SPME fiber was used in combination with ethyl chloroformate for the removal of amino acids in aqueous solutions and aqueous salt solutions. The mechanism for derivatization is shown in Figure 1.
Figure 2 identifies the conceptual locations of the available data sets in the study area; ‘target’ locations correspond to data set locations. Integration of the existing process–driven approach with a data–driven approach involves training an LSTM model to reproduce these ‘target’ data sets. Then, the two models can be integrated so that the process–driven model provides water budget accounting for water movement and storage between data set locations. The use case for this type of integrated model is for historical and future conditions (i.e., periods without data sets).
ACCOMPLISHMENTS
SPME combined with a SwRI developed method using ethyl chloroformate and subsequent analysis via GCMS was successful in the removal, derivatization, separation and identification of amino acids from an aqueous media. Figure 2 shows a representative chromatogram from the SPME injection of a derivatized amino acid aqueous sample.
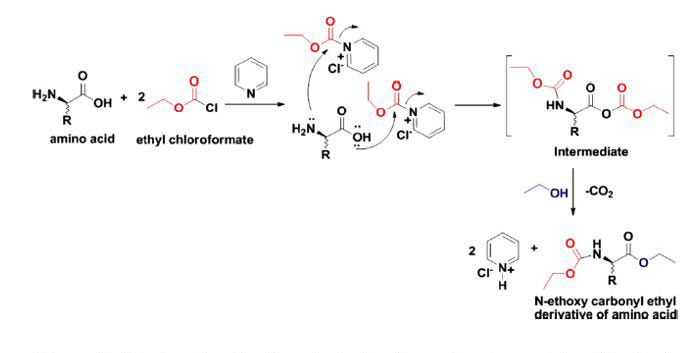
Figure 1: Mechanism for the derivatization of amino acids with ethyl chloroformate.
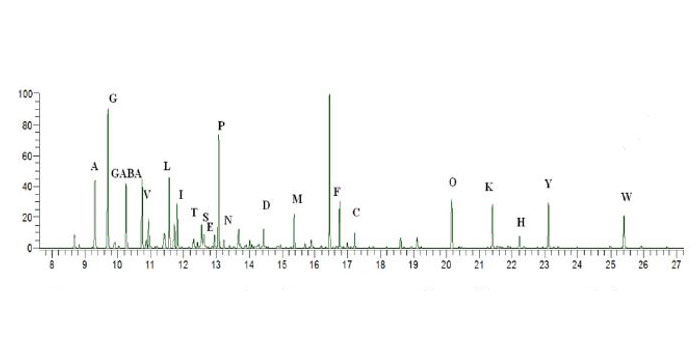
Figure 2. Representative chromatogram from the SPME, ethyl chloroformate derivatization and GCMS analysis of amino acids in a aqueous media.
